How to interpret SMARTCYP results
Explanation of output
- The results are shown in form of a structure and a table for each molecule.
- The 3 top ranking atoms are highlighted both in the structure and table.
- The figure can be switched to showing atom numbers by hovering the mouse pointer over the figure.
The atom numbers will be hidden if you move the pointer away from the figure.
- The table gives the Rank, Atom (type and number), Score, Energy, Accessibility, 2DSASA, and Similarity.
- The atoms in the tables are ranked by Score, with the lowest score resulting in the lowest rank,
and thus the highest probability of being a site of metabolism.
- If the energy is 999, then there is no matching energy rule. Such sites should not be considered as
possible sites of metabolism.
- While the top three sites are coloured for easier identification this does not mean there is
a cutoff saying that there are always three likely sites, the score is what matters.
- The similarity is a number from 0 (low) to 1 (high). It indicates how similar the atom in the molecule is
to an atom in a fragment on which the activation energies have been determined with DFT.
Which model to use?
In the output, there is a table with three tabs: 3A4 , 2D6, and 2C9.
- 3A4 : The standard SMARTCyp score (see below)
was originally benchmarked towards CYP3A4 but performs reasonably well also to
predict sites of metabolism for CYP1A2, 2A6, 2B6, and 2E1.
- 2C9 : The SMARTCyp score 2C9 score was develope for CYP2C9, but is also used to
predict SOMs for 2C8 and 2C19.
- 2D6 : The 2D6 score is used for CYP2D6.
Algorithm for the standard SMARTCyp
Definition of the algorithm used in SMARTCyp and description of the variables in it:
Score, S = E - 8*A - 0.04*SASA
- Energy = E: E is an approximate activation energy for the reaction of the catalytic site of a CYP with
the molecule at this atom. It is decided by fragment matching of each atom against a lookup table with SMARTS rules
and activation energies in kJ/mol.
- Accessibility = A: The accessibility is a relative measure of the topological distance for an atom from
the center of the molecule, and is always a number between 0.5 (atom at the center) and 1 (atom at the end).
- Solvent Accessible Surface Area = SASA: The SASA describes the local accessibility of an atom and is computed using the
2DSASA algorithm which predicts this value from the molecular topology (for an exact value 3D coordinates would be necessary).
Among symmetric atoms only one is in the html output (this works in 99% of all cases, the symmetry perception algorithm is not perfect.
Algorithm for the CYP2D6 model in SMARTCyp
Definition of the CYP2C9 algorithm used in SMARTCyp and description of the variables in it:
Score, S = E + N+dist_correction + Span2End_correction - 0.04*SASA
- N+dist_correction: N+dist is the number of bonds from an atom to a protonated amine. The correction is computed as follows:
- N+dist < 8: N+dist_correction = 6.7 * (8 - N+dist)
- N+dist >= 8: N+dist_correction = 0
- Span2End_correction: Span2End is the number of bonds between an atom and the end of the molecule.
The correction is computed as follows:
- Span2End < 4: Span2End_correction = 6.7 * Span2End
- Span2End >= 4: Span2End_correction = 6.7 * 4 + 0.01*Span2End
Algorithm for the CYP2C9 model in SMARTCyp
Definition of the CYP2C9 algorithm used in SMARTCyp and description of the variables in it:
Score, S = E + COO-dist_correction + Span2End_correction - 0.04*SASA
- COO-dist_correction: COO-dist is the number of bonds from an atom to a carboxylic acid (or a bioisostere thereof).
The correction is computed as follows:
- COO-dist < 8: COO-dist_correction = 5.9 * (8 - COO-dist)
- COO-dist >= 8: COO-dist_correction = 0
- Span2End_correction: Span2End is the number of bonds between an atom and the end of the molecule.
The correction is computed as follows:
- Span2End < 4: Span2End_correction = 5.9 * Span2End
- Span2End >= 4: Span2End_correction = 5.9 * 4 + 0.01*Span2End
Known Limitations
Since the SMARTCyp method relies heavily on reactivity there are some specific types of sites which often are ranked too high or too low.
- Sites with very low 3D accessibility are ranked too high
- Sites which are found as metabolites only due to entropy are ranked too low (for example tertbutyl groups which have
nine identical hydrogen atoms).
- For really large compounds (more than 40 non-hydrogen atoms in CYP3A4) the reactive sites found by SMARTCyp are usually
not the experimentally found metabolites. This is probably because the sites of metabolism for such large compounds depend
heavily on the binding conformation.
Metabolites
SMARTCyp predicts the site of metabolism, but not the metabolite. The following table shows the most common P450 transformations and
can be used to estimate what metabolite could be formed.
Oxidation at aromatic and double bonded carbons can produce a number of different metabolites
such as epoxides, alcohols, diols, ketones etc. There is currently no simple way to determine the most probable
product at such sites, so for these sites the products shown in the table above should be viewed as possibilities.
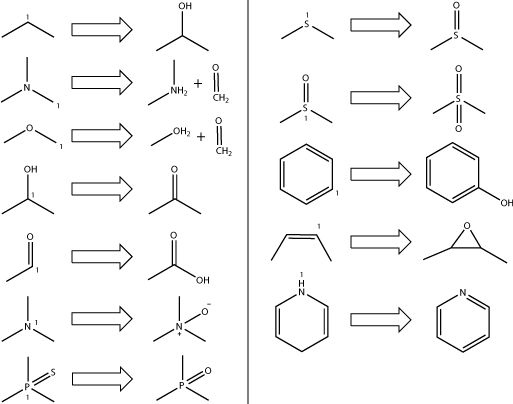 If the atom labelled "1" in the figure above is the predicted site of metabolism, the metabolite formed is
the one showed.
If the atom labelled "1" in the figure above is the predicted site of metabolism, the metabolite formed is
the one showed.
