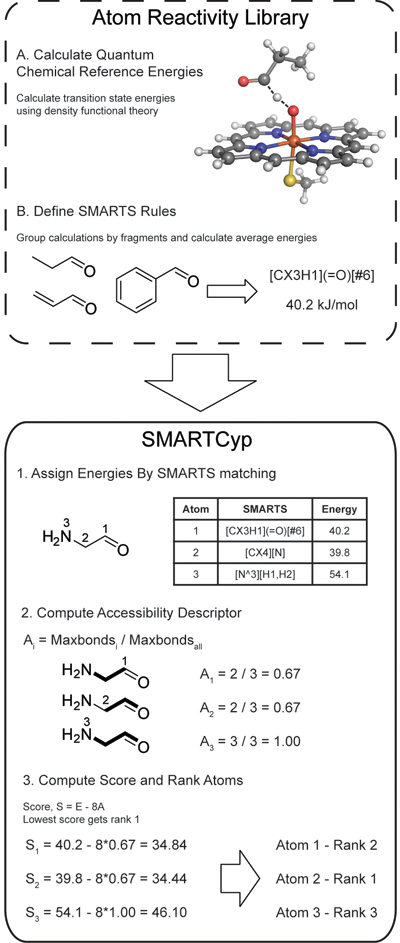
The SMARTCyp method is described in the figure to the right. To construct the program a large number of Density Functional Theory (DFT) calculations of fragment activation energies have been performed. The results have been compiled into rules consisting of SMARTS patterns and associated energies.
SMARTCyp only uses the 2D structure of a compound, and the energy required for oxidation at each atom is computed by fragment matching towards the SMARTS patterns. If multiple patterns match the same atom the pattern with the lowest energy is used. The accessibility is approximated as the relative topological distance of an atom from the center of the molecule, and the final score is computed as Score = energy - 8*accessibility - 0.04*SASA.
The SMARTCyp 2D6 model is slightly more complex, using the basic SMARTCyp reactivity, plus the distance from an atom to the end of the molecule (Span2End), and the distance from an atom to a protonated amine nitrogen atom (N+dist). A linear correction is then applied giving a final 2D6-score = energy + 6.7*(8 - N+dist + Span2End). Cutoffs are applied so that N+dist is never larger than 8, and Span2End is never larger than 4.
The SMARTCyp 2C model is a remake of the 2D6 model, using the basic SMARTCyp reactivity, plus the distance from an atom to the end of the molecule (Span2End), and the distance from an atom to a carboxylic acid (or a bioisoster thereof) oxygen atom (COO-dist). A linear correction is then applied giving a final 2C-score = energy + 5.9*(8 - COO-dist + Span2End) - 0.04*SASA. Cutoffs are applied so that COO-dist is never larger than 8, and Span2End is never larger than 4.
There is also a correction for unlikely N-oxidations. A set of tertiary alkylamines for which the reactivity is a bad representation of the likelyhood for the generation of an N-oxide are given a penalty of 100 kJ/mol to their nitrogen-atom energy. A detailed description of why this is scientifically sound can be seen in the paper where we find that they are actually limited by their N-inversion barrier. (P. Rydberg et al.,Nitrogen Inversion Barriers Affect the N-Oxidation of Tertiary Alkylamines by Cytochromes P450 Angew. Chem, Int. Ed. 2013, 52, 993-997)